General Informations
SNPs&GO is a web server
for the prediction of human disease-related single point protein mutations.
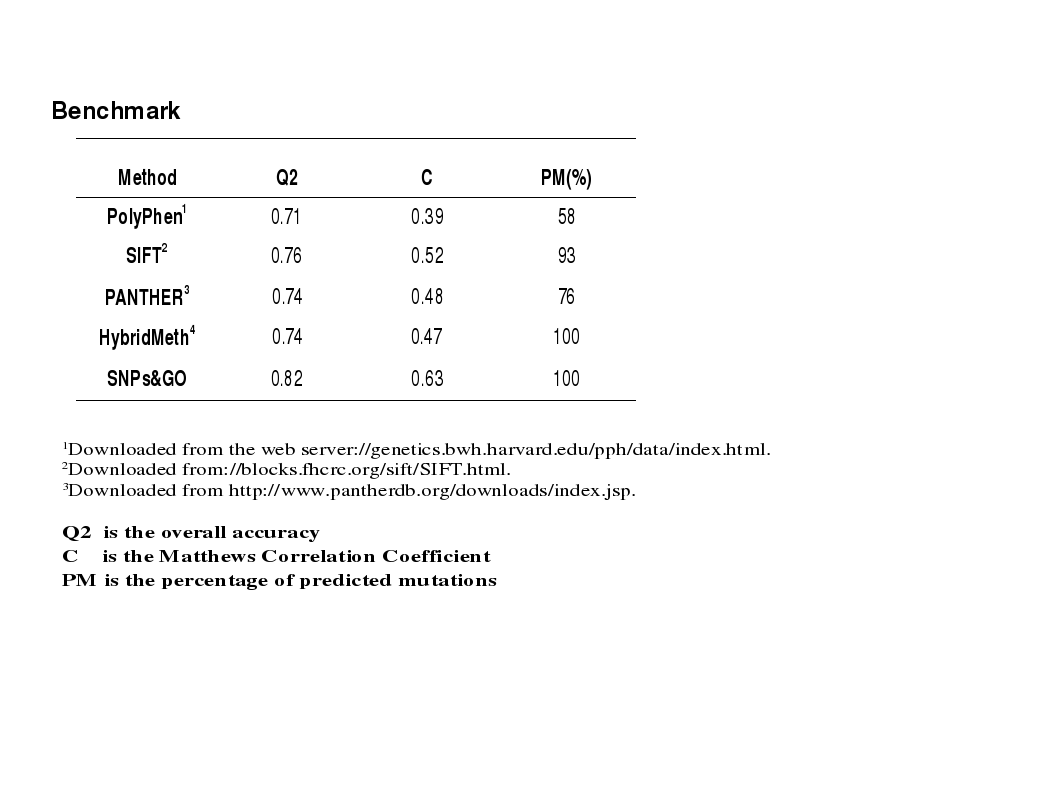
The genetic basis of human variability is mainly due to Single Nucleotide Polymorphisms (SNPs). The most investigated SNPs are missense mutations resulting in residue substitutions in the protein. Here we propose SNPs&GO, an accurate method based on support vector machines, to predict disease related mutations from the protein sequence, scoring with accuracy=82% and Matthews correlation coefficient=0.63. SNPs&GO collects in an unique framework information derived from protein sequence, protein sequence profile, and protein function.
The enormous number of human SNPs available in the data bases poses the question of relating mutations to diseases.
We propose a new SVM based method that uses different pieces of information, including that derived from the Gene
Ontology annotation to predict if a given mutation can be classified disease-related or not. For the first time we
present a GO-integrated predictor tested and trained with a stringent cross-validation procedure.
SNPs&GO was trained on a set of more than 33000 mutations and tested with cross validation procedure over sets in which similar proteins were kept in the same dataset also for the calculation of the LGO score, as derived from the GO data base.
At increasing input level of complexity, the performance is also increasing, suggesting that on top of sequence profile also LGO,
derived from the protein GO annotation, is a crucial added value for discriminating disease related polymorphisms from neutral ones.
The finding that the level of performance increases at increasing information added to the input corroborates the notion that support vector machines can capture all the correlations existing in complementary knowledge.
The benchmark that we performed in house indicates that presently SNPs&GO is one of the best scoring
classifiers available for predicting whether a mutation at the protein level is or is not disease-related.