In order to perform a prediction, the following form has to be filled with:
- a valid UNIPROT Accession Number
- the position of the mutation
- the wild-type residue
- the substituting residue
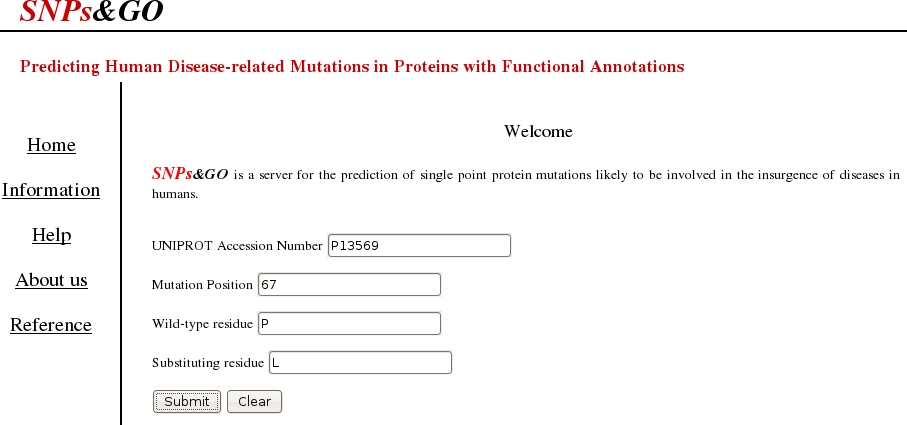
Once filled all the input boxes (All the fields must be filled!)
PRESS the SUBMIT botton to get the prediction output page
OUTPUT

The Output page reports the predicted effect of the mutation uploaded in the prediction form:
- the "EFFECT" column indicates whether the mutation is predicted to be associated to a disease;
- the "RI" column reports the Reliability index of the prediction, scoring from 0 (unreliable) to 10 (reliable).
- the "BP" row reports the links for the Gene Ontology Biological Process terms associated to the protein under investigation and used in the prediction task.
- the "MF" row reports the links for the Gene Ontology Molecular Function terms associated to the protein under
investigation and used in the prediction task.
-the "CC" row reports the links for the Gene Ontology Cellular Component terms associated to the protein under
investigation and used in the prediction task.